Docker for Data Scientists: Simply your workflow and Avoid Pitfalls
Tue 14 August 2018 by Jeff FischerThis is a why-to and how-to guide to using Docker in your data science workflow. It is the companion text to the talk I will be giving at the PyBay Conference in San Francisco on August 18th. Given its length, I am spreading this out over the next week in three posts.
Outline
Here is an outline of the entire series:
Part 1: August 14 (today's post):
Part 2: August 18 (available here):
Part 3: August 21 (available here):
To follow along at home, running code for all the examples is available in a GitHub repository: https://github.com/jfischer/docker-for-data-scientist-examples.
Introduction
These days, DevOps folks live and breathe containers, especially Docker and related technologies. As a Data Scientist, you may have heard about Docker, but are less interested in investing the time to become an expert, since it is not core to your job. If you have tried to learn Docker, you may have even been overwelmed by its complexity and arcane error messages.
Docker can be a very powerful tool and you can learn how to use it without going all the way down the rabbit hole. In this guide, you will get just enough Docker knowledge to improve your data science workflow and avoid common pitfalls.
This guide is based on my experiences as an independent consultant, helping data science teams to introduce reproducible and automated machine learning workflows.
Prerequisites
I assume that you have some familiarity with the command line. The examples are scripted for Linux or Docker for Mac, and will only work out-of-the-box in those environments. The examples could, in theory, be adapted to Docker for Windows, but that is beyond the scope of this guide.
You will need to have a copy of Docker installed. If you do not already have one, you can get the free Community Edition here.
You will also need the code for the examples: https://github.com/jfischer/docker-for-data-scientist-examples. Each subdirectory contains a short description in README.txt, a Dockerfile that describes the container, and a shell script run.sh, that automates the process of building and running the scenario.
What is Docker?
Docker is container platform for deploying isolated applications on Linux. It includes a tool chain for creating, sharing, and building upon layered application stacks. Docker also forms the basis for more advanced services such as Docker Swarm from Docker Inc. and Kubernetes from Google.
Comparing Deployment and Isolation Approaches
To start in our understanding of Docker, we can compare it to other ways of deploying Python applications in an isolated manner.
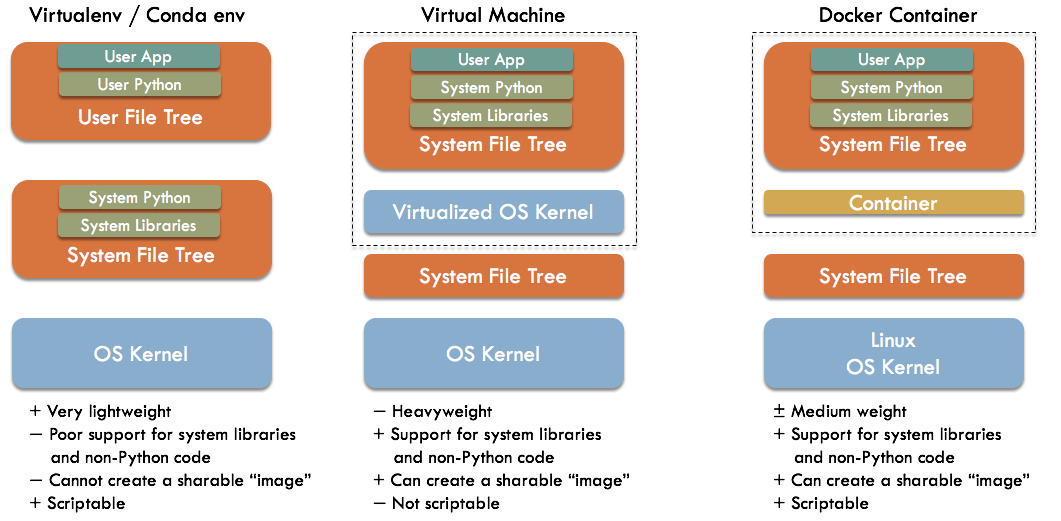
In the figure above, we compare a typical Docker-based stack (on the right) to a stack obtained via the Python tool virtualenv (left) or a virtual-machine-based stack (center).
Virtual environments Virtualenv (Python 2.x) and venv (Python 3.x) are command line tools for creating isolated Python environments. The Anaconda Python distribution and conda package manager have a similar facility. With these tools, a local copy of Python is created under your home directory, as opposed to the primary system file tree where the Operating System's copy of Python typically lives (e.g. under /usr for Linux). This avoids permission problems as well as the issues where you may mess up other programs by making changes to your Python environment. In fact, you can deploy multiple virtual environments within a given user account, isolated from each other. Through the magic of symbolic links to files in the system Python, each environment requires only a small amount of additional storage space.
Overall, this approach is very lightweight and scriptable (since it uses command line tools). However, it provides poor support for installing system libraries and non-Python-based libraries. Also, there is no easy way to create a single sharable file (an "image") that you can just hand to your peer and ask them to run.
Virtual machines Virtualization technology simulates a complete hardware and software stack on top of your existing Operating System. Examples include VMWare (commercial) and VirtualBox (open source). We can create a separate virtual machine for each application we wish to install. The Python application can be installed directly into the operating system's Python tree, since there are no worries about disturbing other applications or users.
Overall, this approach makes it easy to support complex stacks involving non-Python code. You can also create an image file containing the entire virtual machine, which can be shared and started up by your peers. However, virtual machines can take significant system resources. Furthermore, due to long startup times and limited command line support, virtual machine based worflows are not easily scriptable.
Docker Containers Containers make use of Linux kernel facilities to create isolated groups of processes. These processes see their own separate root file trees and can have isolated views of system resources like network ports and users. All the containers appear to the outside system as just regular processes. From an application perspective, you can treat each container almost like a separate machine, installing applications and libraries directly into its root file tree. There are limitations, and you generally cannot run any kind of GUI application.
Overall, containers are much lighter weight than virtual machines (enabling some unique use cases), easily support complex stacks or non-Python code, and can be scripted from the command line. However, containers are more complex and more heavyweight than using a Python virtual environment.
As a Data Scientist, Why Should You Care?
Automation of deployment via Docker containers helps you to focus on your work, and not on maintaining complex software dependencies. By freezing the exact state of a deployed system inside an image, you also get easier reproducibility of your work and collaboration with your colleagues. Finally, you can use resources like Docker Hub to find pre-built recipes (Dockerfiles) from others that you can copy and build on.
Automating Data Science Workflows
The rest of this guide is organized around four representative workflows that we will run using containers:
- Run a Python script
- Run a development session
- Run a Jupyter notebook
- Load and run a database
For each workflow, we will look at how we can implement it using Docker, see example code, and see what happens when we run it. As mentioned above, all of the example code is available in a GitHub repository: https://github.com/jfischer/docker-for-data-scientist-examples. The code should run as-is on Linux or using the MacOS command line and Docker for Mac.
Next Post: Part 2
Thanks for reading part 1 of this series. Part 2 is now available! To continue reading, click here.