How to Build a Reproducible Data Science Workflow
Thu 19 April 2018 by Jeff FischerWhy care about reproducibility? You probably have heard about reproducibility in the context of scientific experiments. It is about providing enough information to the readers of a publication to enable them to run the same experiment and see the same results. This includes information about experimental procedures, materials used, and, increasingly, software.
In machine learning projects, one is frequently running mini-experiments to see the impact of feature or (hyper)parameter changes on a given model. Even if you never publish a scientific paper, reproducibility can be important for these types of experiments. Here are a few reasons why:
- You want to systematically explore all reasonable parameter combinations and understand which ones have an impact on the results.
- You might obtain a result, but get a different one a few days later. Is that because you misremembered the parameters, a change to the code/data, or just random variance?
- You want to move a given model to production, and run it across hundreds of computers in the cloud. How do you ensure consistent results?
- You shelve a project. A few months later, you want to take it in a new direction. Can you pick up where you left off? Even worse, your company wants to revive a project, but the team moved on years ago. Can you even find the original data? [Been there, done that!]
Reproducibility and the Data Pipeline
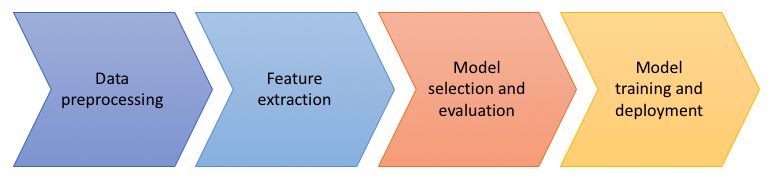
Let us look at reproducibility from the perspective of a data pipeline. A typical machine learning project might have a pipeline with four overall stages:
- Data preprocessing - get the data from its original form into something that can easily be worked with and perhaps combine data from multiple sources.
- Feature extraction - convert a subset of the data into numeric form to be used as inputs to the training. This may involve various kinds of aggregations and encodings.
- Model selection and evaluation - try out various model and solver parameter combinations and see which perform the best against some kind of scoring function.
- Model training and deployment - after selecting and validating a model, train it and deploy it to make classifications or predictions.
Note that there are data dependencies introduced across each of these stages. If I change a (hyper)parameter in stage three, I have to rerun the training in stage four. If I change something in the data preprocessing stage, I will need to rerun all the subsequent stages. In order to keep our sanity, we would want to change only one thing at a time. That means subsequent stages should run the same way each time, with the only changes being in their inputs.
Categorizing Data
How do we address this problem? Let us start by breaking it down into smaller subproblems. We will consider four types of data:
- Source data is the raw data set used as input. For machine learning, you can never have enough data, and your source data is like gold. You need to protect it and keep track of any changes.
- Code that you write to implement data transformations and machine learning algorithms.
- Configuration that includes the software environment in which your pipeline is executing, with all the third party dependencies.
- Intermediate data and experimental results that you produce as outputs of each stage of the pipeline.
Now, let us look at some solutions for each data category in more detail.
Source Data
To protect source data, you need a well-defined place to keep it, easy backups or copies, a way to version individual data items, and a way to tag and retrieve a collection of data items. For the underlying infrastructure, here are some commonly-used options:
- Source Code Repositories like git are a good place for source data. You can keep the repository on a central server and easily make copies to developer desktops or compute resources by checking out the repository. All files are versioned and you can tag the repository, allowing you to retrieve the set of files at a point in time or associated with a milestone in your project. Scaling, or lack thereof, is the big disadvantage of source control repositories. Github has hard limits on file sizes and commit sizes. Even without hard limits, you will run into performance issues with large git repositories.
- Object Storage such as Amazon's S3 service can store a hierarchy of files without the size limitations of source code repositories. S3 also supports versioning, which lets you easily keep and retrieve the older versions of your data. Although S3 provides the ability to store metadata (key/value pairs) with each object, there is no concept of associating a tag with a collection of files and then later retrieving those specific versions of the files. Something like this could certainly be built on top of the primitives S3 provides.
- File Servers can be used within an intranet for storing source data. These can scale to large data sizes and use standard protocols like SMB and NFS to permit shared access. Unfortunately, standard filesystems do not support versioning of individual files. Some filesystems and file servers support snapshots which allow you to cheaply keep multiple versions of the entire file tree. However, these are usually only available to the system administrator and not as lightweight as a git version tag. Thus, people often resort to ad hoc approaches that need a lot of discipline to follow.
Independent of your choice of infrastructure, you need to keep documentation about where you got the data from, what it contains, and the format of the files. It would be unfortunate if you come back to the data set a few years later and find a collection of binary files ending in .dat with no hint to the data format.
The documentation needs to be kept and versioned with the data, so that it is not later lost.
Code
Code is the most straightforward of these -- you should always be using a source control system. All software artifacts, including build scripts and application metadata should be kept under source control. You also have to be in the habit of to checking in your files before running an experiment. You can use commit messages and tagging to associate versions of your software with specific experiments and data versions.
Configuration
The stack of third party software used to support your experiments needs to be tracked as well. This includes programming languages, data format parsers, databases, domain-specific libraries, and machine learning frameworks. A modern application may easily involve a hundred components or more, managed by multiple tools (e.g. the OS package manager, the language package manager, and just hand-installed components).
Containers
To manage all this complexity without having to hand-track all the details, I suggest using Docker containers. Containers are lightweight components similar to virtual machines that encapsulate the full root filesystem of a system environment (including all system utilities, installed software packages, and user data files) and provide a level of resource isolation.
(Docker) containers have two key advantages over other technologies:
- They can be layered -- you can take an existing container specification (say of Ubuntu Linux with TensorFlow installed) and add on you own components.
- These layers are specified in a straightforward text-based format called a Dockerfile, which can be stored and versioned in a source code repository along with your code. This makes it easy to reproduce a given container and to tie specific container versions with versions of your code.
You can and should store pre-compiled Dockerfiles ("images") for performance reasons and in case a third-party dependency is no longer available. However, you always have the option to reproduce a container from the source Dockerfile. If you treat containers as immutable, throwaway services, you can avoid the problems that occur with virtual or physical machines when people manually make changes without religiously documenting their actions. For that reason, you should never store results or derived data in your containers.
Application Parameters
Not all of your software configuration is easily expressed in a Dockerfile. Most data science experiments have fine-grained parameters that may control the features generated, scaling factors, solver strategies, and regularization. For these, you might consider including them in configuration files that are managed by your source control repository or as command line parameters. In the second case, you should record the parameters along with the results you generate.
Intermediate Data and Results
Like source data, some common solutions for intermediate and result data storage include code repositories, object storage, and file servers. Storing intermediate data and results does have different trade-offs from the source data. It usually is smaller than source data, so it does not necessarily have scaling issues. However, this kind of derived data is frequently changing, so tracking the versions and tying the results to how they were produced becomes very important.
I am not aware of an existing open source or commercial tool which solves the reproducibility problem for this data without requiring you to adopt someone's end-to-end orchestration infrastructure. I can recommend the following best practice:
Whenever your software generates a data output, information about the pipeline which generated the data should be saved along with it.
This means tying the derived data to the other three types of data: the source data used as the initial input, the version of your source code, and the software configuration. Let us look at each of these individually:
- For source data, you should have metadata about the version and source of the data that is copied along the data pipeline.
- For source code, you can record the version of the source repository. For example, git has a command git describe --tags which will output the most recent tag's name, the number of commits since the last tag was taken, and the hash of the current repository.
- For containers, you can version the Dockerfiles along with your source code. You can also track and record the label you associate with each image (a name and version).
- Command line parameters can be written out with the results.
By doing this, you will always know the lineage of your derived data and results.
Conclusion
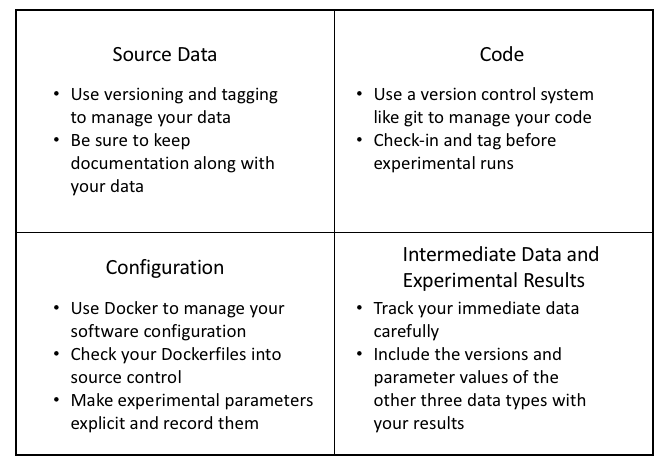
Summary of Best Practices for Data Science Reproducibility
In this blog post, we have seen the reasons why you should care about reproducibility, even if you are not working toward a scientific publication. We have also seen the role of the data pipeline in the production of machine learning data and results. Finally, we looked in detail at four key types of data you need to manage and some options and best practices specific to each data type. The figure above summarizes these best practices.
I hope that you found this useful. If you have thoughts about this, reach out to me and let's talk!